University of Illinois Urbana-Champaign
Biomedical Optical Imaging Laboratory
Overview
The laboratory of Dr. Yang Liu focuses on developing multiscale multimodal optical microscopy techniques spanning seven orders of magnitude, bridging the nanoscale to the mesoscale, to significantly advance precision medicine such as early detection, prevention, and treatment of cancer. The technologies integrate label-free quantitative phase imaging with hyper-plex mesoscale microscopy, super-resolution fluorescence microscopy, and highly sensitive and specific across-the-scale imaging probes, which are paired with artificial intelligence (AI), robotic automation, and large-scale image informatics. This transformative approach allows for dynamic high-content phenotyping and provides invaluable insights into the molecular changes under complex tissue microenvironments. This is critical in understanding the progression from precursors to invasive cancer and deciphering the mechanisms behind therapeutic resistance. The fusion of cross-scale imaging with AI-driven systems biology and automation is poised to drive future scientific discoveries and transform personalized medicine. Here is a recent poster summarizing our research.
Technology Development
Multiscale multimodal hyperplexed imaging platform for spatiotemporal biology
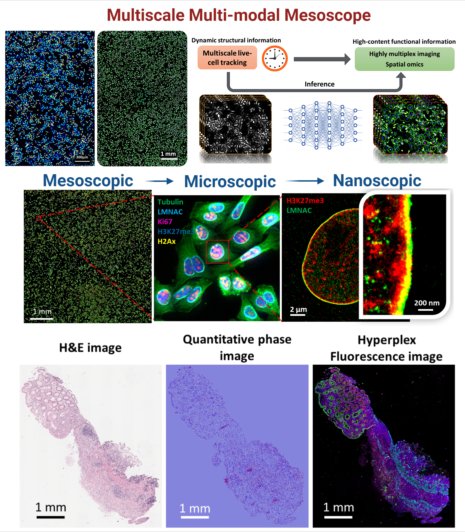
Spatial biology offers transformative insights into cellular function within microenvironments, pivotal for precision medicine. Our multiscale multimodal imaging platform is tailored for quantitative spatiotemporal biology. This platform seamlessly integrates quantitative phase, hyper-plex fluorescence imaging, and a wide field-of-view with sub-micron resolution. It also co-registers mesoscale images with molecular-scale super-resolution images and whole-slide histology images, bridging molecular information with mesoscale cell sociology and tissue contexts. This platform will serve as a powerful tool to enhance spatial biology by allowing rapid and comprehensive profiling of mesoscale microenvironment and spatial heterogeneity, temporal dynamics of cell morphology and their underlying functional and molecular characteristics over a large cell population in cells under different treatments and tissue at various pathological conditions.
Relevant Publcations:
1. Cang H, Liu Y, Xing J. Mosaic-PICASSO: accurate crosstalk removal for multiplex fluorescence imaging.
Bioinformatics, 40 (1): btad784, 2024.
2. Ma H, Chen M, Xu J, Yang Y, Zhao Y, and Liu Y. An Omni-Mesoscope for multiscale high-throughput quantitative phase imaging of cellular dynamics and high-content molecular characterization.
Science Advances, 10, eadq5009, 2024
3. Chen M, Ma H, Sun X, Schwartz M, Brand RE, Xu J, Gotsis DS, Nguyen P, Moore BA, Snyder L, Brand RM, Liu Y. Multimodal whole slide image processing pipeline for quantitative mapping of tissue architecture and tissue microenvironment.
npj Imaging, 3, 26, 2025.
High-throughput STORM
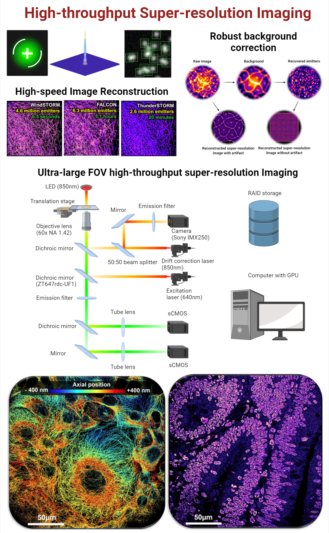
Advances in super-resolution fluorescence microscopy have revolutionized biological imaging by overcoming the fundamental diffraction barrier, recognized by a Nobel Prize in 2014. In particular, single molecule localization microscopy (SMLM) such as (fluorescence) photo-activated localization microscopy [(f)PALM] and (direct) stochastic optical reconstruction microscopy [(d)STORM] is one of the simplest and most cost-effective approaches to achieve super-resolved imaging capability at a spatial resolution down to 5-10 nm. Despite its impact, it is generally deemed as a complex and low-throughput technique. The main rate-limiting factors are the small illumination field, the need to acquire tens of thousands of imaging frames to reconstruct an image and slow image reconstruction speed. It is also limited to imaging thin and transparent samples (bacteria, cultured cells). We aim to develop multi-color high-throughput STORM system that can be used to analyze clinically relevant pathological samples and apply machine-learning tools for automated analysis.
Relevant Publcations:
1. [Xu J#, Ma HQ#], Ma HB, Jiang W, Duan M, Mela CA, Zhao S, Gao C, Hahm E-R, Lardo SM, Troy K, Sun M, Pai R, Stolz DB, Zhang L, Singh S, Brand RE,
Hartman DJ, Hu J, Hainer SJ, Liu Y. Super-resolution imaging reveals the evolution of higher-order chromatin folding in early carcinogenesis.
Nature Communications, 11: 1899, 2020.
2. Ma H, Xu J, Liu Y. WindSTORM: Robust online image processing for high-throughput nanoscopy.
Science Advances, eaaw0683, 2019.
3. Mela CA, Liu Y. Application of convolutional neural networks towards nuclei segmentation in localization-based super-resolution fluorescence microscopy images.
BMC Bioinformatics, 22, 325, 2021.
4. Xu J, Sun X, Kim K, Brand RM, Hartman D, Ma H, Brand RE, Bai M, Liu Y. Ultrastructural visualization of chromatin in cancer pathogenesis using a simple small-molecule fluorescent probe.
Science Advances, 8: eabm8293, 2022.
5. Xu J, Ma H, Liu Y. Stochastic optical reconstruction microscopy (STORM). Current Protocols in Cytometry, 81: 12.46.1–12.46.27, 2017.
6. Ma H, Chen M, Nguyen P, Liu Y. Towards drift-free high-throughput nanoscopy through adaptive intersection maximization. Science Advances, 10(21): 1239-1246, 2024.
7. Ma H, Nguyen P, Liu Y. A Versatile Drift-Free Super-Resolution Imaging Method via Oblique Bright-Field Correlation. Advanced Science, 12, 2412127, 2025.
Multiscale Quantatitative Phase Imaging
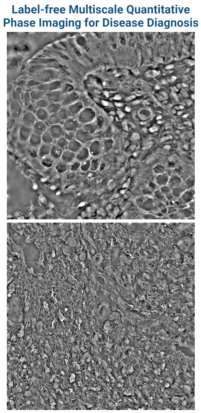
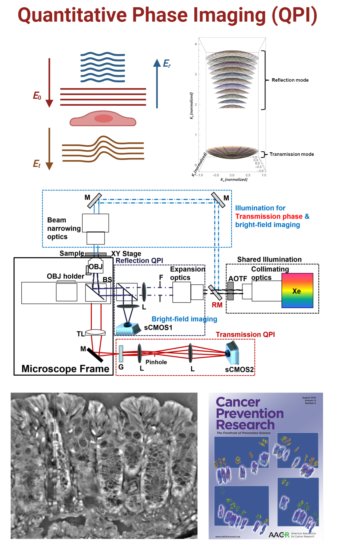
Quantitative Phase Imaging (QPI) offers a label-free approach to non-invasively characterize cellular processes by exploiting their refractive index-based intrinsic contrast. QPI captures this contrast by translating refractive index-associated phase shifts into intensity-based quantifiable data with nanoscale sensitivity. The integration of spatial biology, multi-omics, and artificial intelligence with QPI’s precise and unbiased mapping of tissue architecture and nanoscale details could serve as the next-generation approach in advancing precision medicine through label-free disease diagnosis, monitoring, and treatment.
Relevant Publcations:
1. Pham HV, Pantanowitz L, Liu Y. Quantitative phase imaging to improve the diagnostic accuracy of urine cytology.
Cancer Cytopathology, 124:641-650, 2016.
2. Liu Y, Uttam S. Perspective on Quantitative Phase Imaging to Improve Precision Cancer Medicine.
Journal of Biomedical Optics, 29 (S2): S22705, 2024.
3. Ma H, Chen M, Xu J, Yang Y, Zhao Y, and Liu Y. An Omni-Mesoscope for multiscale high-throughput quantitative phase imaging of cellular dynamics and high-content molecular characterization.
Science Advances, 10, eadq5009, 2024
Nanoscale nuclear architecture mapping and QPI
Nanoscale nuclear architecture mapping (nanoNAM) is an interferometry approach to capture the sub-resolution nanoscale structural alterations within the cell nucleus with slowly varying refractive index in the backcattered signals. Based on our Fourier mapping approach, nanoNAM computes the Fourier phase associated with these back-scattered waves to capture, with nanoscale sensitivity, (a) depth-resolved alterations in mean optical density, and (b) depth-resolved localized heterogeneity in optical density of the cell nuclei. This depth-resolved capability of nanoNAM allows 3D quantification of nuclear architecture, while simultaneously avoiding the confounding effect of thickness variation. We apply nanoNAM together with high-contrast quantitative phase imaging to explore its ability to detect subtle structural changes in refractive index in cells undergoing early-stage carcinogenesis.
Relevant Publcations:
1. [Uttam S*, Pham HV*], LaFace J, Leibowitz B, Yu J, Brand RE, Hartman DJ, Liu Y. Early prediction of cancer progression by depth-resolved nanoscale maps of nuclear architecture from unstained tissue specimens.
Cancer Research, 75(22): 4718-4727, 2015.
2. Uttam S, Hashash JG, LaFace J, Binion D, Regueiro M, Hartman DJ, Brand RE, Liu Y. Three-Dimensional Nanoscale Nuclear Architecture Mapping of Rectal Biopsies Detects Colorectal Neoplasia in Patients with Inflammatory Bowel Disease.
Cancer Prevention Resesarch, 12(8), 527-538, 2019.
3. Uttam S, Liu Y. Fourier phase based depth-resolved nanoscale nuclear architecture mapping for cancer detection.
Methods, 136: 134-151, 2018.
4. Uttam S, Liu Y. Fourier phase in Fourier-domain optical coherence tomography.
Journal of the Optical Society of America A, 32(12):2286-2306, 2015.
5. Thota PN, Nasibli J, Kumar P, Sanaka MR, Chak A, Zhang X, Liu X, Uttam S* and Liu Y*. Prediction of Neoplastic Progression in Barrett’s Esophagus Using Nanoscale Nuclear Architecture Mapping: A Pilot Study.
Gastrointestinal Endoscopy, 95(6): 1239-1246, 2022.
miniSTORM

Although super-resolution microscopy is commercially available, it remains a high-end microscopy instrument only available in the core imaging centers at major academic institutions and a small number of well-funded laboratories around the world. The high cost of this technology limits its widespread use as a routine microscopy system. Small colleges and commercial laboratories often cannot afford such expensive instruments. We aim to develop miniSTORM, an affordable state-of-the-art nanoscopy with a spatial resolution one order-of-magnitude better than conventional light microscopy. The hardware cost will be 10 times lower than commercial systems. It is compact and can be put on a regular table top without a dedicated room and high-end optical table. It is highly robust and automated. Importantly, miniSTORM integrates artifact minimization modules that provide high-quality image, it will reduce the learning curve and the need for significant user expertise.
Relevant Publcations:
1. Ma H, Liu Y. Robust emitter localization with enhanced harmonic analysis. Optics Letters, 46, 5798-5801, 2021.
2. Ma H, Liu Y. Embedded nanometer position tracking based on enhanced phasor analysis. Optics Letters, 46, 3825-3828, 2021.
3. Ma H, Fu R. Xu J, Liu Y. A simple and cost-effective setup for super-resolution localization microscopy. Scientific Reports, 7(1): 1542, 2017.
4. Ma H, Xu J, Jin J, Huang Y, Liu Y. A simple marker-assisted three-dimensional nanometer drift correction method for super-resolution microscopy.
Biophysical Journal, 112: 2196-2208, 2017.
Light-sheet Microscopy

Light-sheet microscopy is a powerful approach to minimize photobleaching effect and reduce the out-of-focus background. However, the physical constraint of the objective limits the use of high-NA optics, thus limiting the optical resolution. Significant progress has been made in the development of high-resolution light-sheet systems. Our group aims to develop simple high-resolution light-sheet microscopy systems for 3D live-cell imaging for various biomedical applications.
Biomedical Applications
Cancer Detection
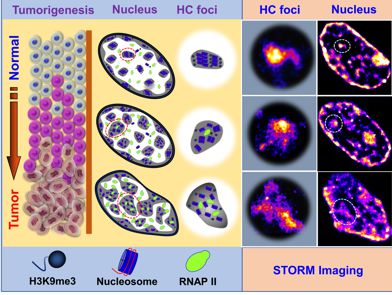
Cancer risk stratification—identifying subjects at high risk for developing cancer—is an important tool in cancer prevention and early detection, and crucial for reducing cancer-related morbidity and mortality. However, due to the immense molecular heterogeneity at each stage of carcinogenesis, few tools are available to accurately identify the small percentage of subjects at the highest risk for developing cancer and who would benefit most from early detection and cancer prevention strategies. Higher-risk pre-cancerous lesions destined to undergo progression to cancer may be indistinguishable from low-risk precursors. Uncertain cancer risk can lead to sub-optimal clinical management of at-risk patients: high-risk patients may undergo inadequate surveillance, whereas the lower-risk subjects may undergo excessive testing or unnecessary interventions.
Abnormal chromatin structure is one of the most universal and striking characteristics in cancer cells and has remained the gold standard for cancer diagnosis for two centuries, but it often lacks sufficient sensitivity to accurately identify patients at highest risk for developing cancer, and cells early carcinogenesis may not appear pathologically distinct. Further, tumor microenvironment (TME) also plays an indispensable role in the disease progression. Comprised of a dynamic interplay among emerging tumor cells, local stromal cells, the extracellular matrix, and signaling molecules, the TME in precursors can provide early cues about the potential for a lesion to transition towards malignancy. Current technology does not offer a comprehensive view of chromatin abnormalities in the context of the tumor microenvironment.
We aim to apply our multi-scale multi-modal imaging into cancer risk stratification, by detecting the cumulative disruption in chromatin folding coupled with their tumor microenvironment to characterize the risk of precursors progressing to malignancy. We are applying these techniques in cancer risk stratification of colon polyps, early detection of lung cancer and detection of aggressive prostate cancer.
Relevant Publcations:
1. [Xu J#, Ma HQ#], Ma HB, Jiang W, Duan M, Mela CA, Zhao S, Gao C, Hahm E-R, Lardo SM, Troy K, Sun M, Pai R, Stolz DB, Zhang L, Singh S, Brand RE,
Hartman DJ, Hu J, Hainer SJ, Liu Y. Super-resolution imaging reveals the evolution of higher-order chromatin folding in early carcinogenesis.
Nature Communications, 11: 1899, 2020.
2. Xu J, Sun X, Kim K, Brand RM, Hartman D, Ma H, Brand RE, Bai M, Liu Y. Ultrastructural visualization of chromatin in cancer pathogenesis
using a simple small-molecule fluorescent probe. Science Advances, 8: eabm8293, 2022.
3. Xu J, Sun X, Chen Z, Ma H, Liu Y. Super-resolution Imaging of T Lymphocyte Activation Reveals Chromatin Decondensation and Disrupted Nuclear Envelope.
Communucations Biology, 7: 717, 2024.
Drug resistance and screening
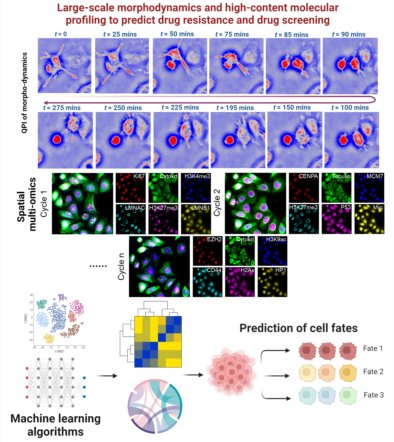
Cancer cells respond to a therapeutic agent and develop resistance over time with various phenotypic representations. The phenotype of an individual cell is determined by its functional state, abundance, morphology, and dynamic behavior of its organelles. Therefore, it is important to understand such diverse phenotypic transitions for a large cell population over the duration of drug treatment. Traditional techniques based on high-content microscopy generally capture a snapshot of the dynamic process, while missing the critical information about the cell morphologic changes in live cells. On the other hand, label-free live-cell imaging captures the dynamic changes of cellular morphology but missing high-content molecular information. To understand the molecular characteristics of morphological dynamics, we have developed a unique multi-scale multimodal mesoscopic imaging system that is capable of label-free quantitative phase imaging of live cells for real-time dynamic tracking of morphological features and hyperplex high-content fluorescence imaging across a wide-range of spatial scales from mesoscale to nanoscale. Leveraging this powerful platform, we aim to identify factors contributing to resistance in cancer treatments and develop more effective therapeutic approaches.
Chromatin Biology
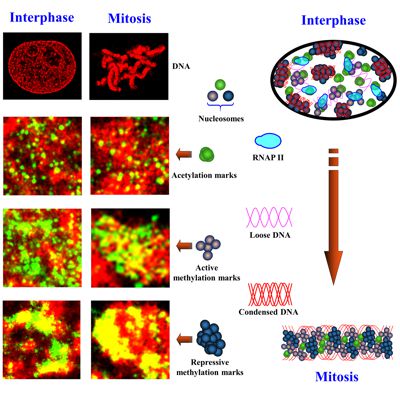
Eukaryotic cells package genomic DNA up to 2 meters long into a nucleus with a diameter of several microns through a hierarchical scheme of compaction into DNA-protein assemblies. The first level is nucleosome, consisting of 147 base pairs of DNA wrapped around an octamer of four core histone proteins. This basic repeating unit of nucleosomes is then organized into ~10 nm “beads-on-string” chromatin fiber, which is further compacted into higher-order chromatin structure, to fit into the micron-sized nucleus. Chromatin organization is regulated by a large number of chemical modifications, particularly on the N-terminal tails of histone core proteins, such as acetylation and methylation. Histone modifications regulate the packaging of nucleosomes into higher-order chromatin structure to influence the accessibility of genomic DNA to the transcription machinery proteins. Subsequently, chromatin compaction at different epigenomic states controls their gene expression and imposes a significant effect on many cellular processes, such as DNA replication, cell division, DNA damage and DNA repair. We apply powerful super-resolution microscopy and genomic technologies (ChIP-seq and RNA-seq) to study higher-order chromatin organization at differnet epigenomic states and aim to understand how chromatin organization coordinates with co-factors to impact gene expression in normal cellular processes and pathological states.
Relevant Publcations:
1. Xu J, Ma H, Jin J, Uttam S, Fu R, Huang Y, Liu Y. Super-resolution imaging of higher-order chromatin structures at different epigenomic states in single mammalian cells.
Cell Reports, 24: 873-882, 2018.
2. Han L, Mich-Basso JD, Li Y, Ammanamanchi N, Xu J, Bargaje AP, Liu H, Wu L, Jeong J-H, Franks J, Stolz DB, Wu Y, Rajasundaram D, Liu Y, Kühn B. Changes in nuclear pore numbers control nuclear import and stress response of mouse hearts.
Developmental Cell, 57(20): 2397-2411, 2022.
3. Xu J, Liu Y; Probing chromatin compaction and its epigenetic states in situ with single-molecule localization-based super-resolution microscopy.
Frontiers in Cell and Developmental Biology, 2021; 9, 653077.
4. Xu J, Liu Y. A guide to visualizing spatial epigenomics with super-resolution microscopy.
FEBS Journal, 2019, 286(16): 3095-3109.
Aging
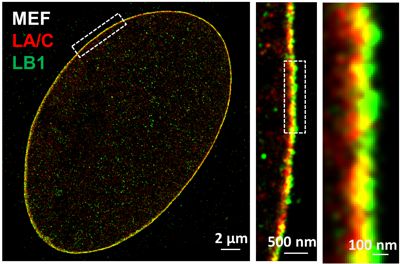
The nuclear lamina is an intermediate filament meshwork adjacent to the inner nuclear membrane that plays a critical role in maintaining nuclear shape and regulating gene expression through chromatin interactions. In collaboration with Dr. Quasar Padiath from Department of Human Genetics at University of Pittsburgh, Dr. Kris Dahl from Carnegie Mellon University and Dr. Bernhard Kuhn from Chilren's Hospital of Pittsburgh, we apply multi-color super-resolution microscopy to characterize distinct ultra-structures of nuclear lamina proteins and understand their functional roles in normal cellular processes and aging.
Relevant Publcations:
[Nmezi B#, Xu J#, Fu R#], Armiger T, Bey G, Schneider J, Ma H, Tu Y, Chen N, Young S, [Dahl K*, Liu Y*, Padiath Q*]. Concentric organization of A- and B-type lamins predicts their distinct roles in the spatial organization and stability of the nuclear lamina.
Proceedings of the National Academy of Sciences of the USA, 116(10): 4307-4315, 2019.